Have you followed the Go contest between Google AlphaGo and the world champion Ke Jie, or heard the news that the medical robot Watson travelled to China and issued a diagnosis and prescription for cancer patients in only ten seconds? What about the ability of artificial intelligence to outperform professional pathologists on recognising breast cancer?
In recent years, with the rapid development of big data and artificial intelligence, and the encouragement from the Chinese government of the advances in the healthcare industry and medical artificial intelligence (AI), it is perhaps inevitable that digital medicine will become the new norm. Utilising AI technology, the government may save more budget in order to more effectively fund medical supplies for disease management and nursing services.
Two Major Research Directions
Aside from digital medicine, as novel and better techniques for biological analysis emerge, the quantity of medical data grows tremendously. Using big data, we can solve problems that were formerly unsolvable. Big data technologies and applications in bioinformatics include personal genome sequencing using high- throughput sequencing, transcriptomic and proteomic research, genotypic and phenotypic studies at single-cell level, metagenomics in human health, and medical image processing. Deriving hypotheses and mathematical models from the knowledge and observed patterns of biological systems to analyse and interpret data is the fundamental initiative and goal of using big data in bioinformatics.
Established in 2016, Prof Douglas Zhang’s laboratory at the University of Macau (UM) focuses on two areas: 1) Continuous monitoring of human physiological signals with wearable devices, which in turn provides data for studying the relationship between physiological dynamics and diseases such as diabetes, respiratory and cardiac disorders; and 2) Genomic, transcriptomic and proteomic studies using high- throughput sequencing, from which the correlation between precision medicine and related diseases can be investigated. This research has been actively supported by the government of Macao, the Science and Technology Development Fund (FDCT), UM, and the Faculty of Health Sciences (FHS).
Digital Health
Diabetes is a group of chronic metabolic disorders. It occurs mainly due to the pancreas not producing enough insulin or the cells of the body not responding properly to the insulin produced. Finally, the body’s blood glucose rises to a high level. China has the world’s largest number of patients with diabetes. According to national surveys, the incidence of diabetes in China has increased dramatically over the past 30 years. Older people, men, residents of cities or economically developed areas, and overweight or obese people have a higher prevalence of diabetes. How can we monitor glucose in a convenient and quick way? How might we prevent the occurrence of diabetes? How is it possible to analyse and process the continuously monitored data and explore information from the massive data? These are the problems that Prof Zhang’s laboratory studies.
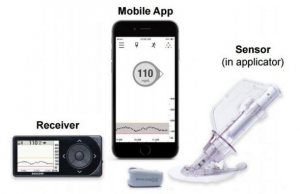
Currently, patients with diabetes monitor their blood glucose by pricking their fingers in the hospital or at home. Generally, they need to prick their fingers with a small needle no more than four times to test the blood glucose levels. The testing method is painful and inconvenient, which may cause low frequency of glucose monitoring and even poor glucose control. For those patients in a pre-diabetes state, the continuous monitoring of glucose can guide the provision of advance interventions and help to restore health. In a cooperative project with the Department of Endocrinology in the First Affiliated Hospital of Guangzhou Medical University, Prof Zhang explored the complexity and fractality of glucose dynamics in patients with type-1 diabetes, type-2 diabetes, pre-diabetes, and normal health. The subjects applied minimally invasive wearable devices (see Figure 1) to continuously monitor the blood glucose. Prof Zhang also developed a R package and related functions to analyse and compare the complexity and fractality.
In this project, Prof Zhang developed an R package CGM analyzer and he found that there was differentiation of complexity and fractality among type-1 diabetes, type-2 diabetes, prediabetes and health subjects (Figure 2). These results prompted Prof Zhang to conduct research to explore the use of complexity and fractality to assess the conditions of patients with prediabetes and then help doctors to treat the patients and adjust their life style, diet, and exercise. The developed R package contains functions for analysing data from a CGM study. It covers a wide and comprehensive range of data analysis methods including reading a series of datasets, obtaining summary statistics of glucose levels, plotting data, transforming the time stamp format, fixing missing values, evaluating the mean of daily difference and continuous overlapping net glycaemic action, calculating multiscale sample entropy, conducting pairwise comparison, and displaying results using various plots. This package is published in the journal Bioinformatics. This package has greatly facilitated the analysis of various CGM studies and helped scientists conduct research about diabetes.
In addition to cooperation with hospitals, Prof Zhang’s laboratory has conducted an experiment in Macao to explore the effects of exercise, heart rate, sleep and diet on healthy people and diabetics. In the experiment, they used Fitbit charge 2 device to monitor the heart rate, sleep quality and exercise status, while the glucose fluctuations were continuously monitored by using the Freestyle blood glucose meter, and the diet was recorded and photographed by the subjects themselves. The research attracted many participants. The project is currently in the process of data analysis.
Prof Zhang also uses wearable medical devices to conduct studies on cardiovascular diseases and respiratory diseases such as chronic obstructive pulmonary disease, asthma and allergic rhinitis.
Bioinformatics
Prof Zhang’s laboratory has also initiated several research projects on bioinformatics. In the integrative transcriptomic analysis on the in-depth exploration of the mechanisms of allergic bronchopulmonary aspergillosis (ABPA), we utilise next-generation sequencing to study respiratory disease-related non-coding RNA, such as long non-coding RNA (lncRNA) and circular RNA (circRNA). We have collaborated with Guangzhou Medical University First Affiliated Hospital and Guangzhou Respiratory Tract Disease Research Centre. Through comparative transcriptomic studies between healthy individuals and patients, we discover disease-related non-coding RNA, which acts as candidates of biomarkers for molecular diagnosis as well as targets in drug design. Moreover, collaborating with Prof Ge Wei in the FHS, employing zebrafish with different mutations (eg YX-1 gene knockout with CRISPR/Cas9), we perform transcriptomic analysis to explore gene-related lncRNA, which helps clarify the regulation of the gene and thus provides the molecular mechanism of the specific gene during zebrafish development.
Through collaboration with Prof Chuxia Deng in the FHS, we investigate the classification and differentiation of breast tumour cells with rats as model using the new high-throughput RNA sequencing technique, Drop-seq. Deng’s laboratory carries out the bioassays and RNA sequencing to obtain Drop-seq dataset, and our laboratory performs data analysis, including expression quantification, cluster analysis, and marker gene identification, to explore the classification and possible molecular mechanisms underlying differentiation of breast tumour cells of rats.
In the project of library construction of whole genome and high-throughput screening based on CRISPR/Cas9, Zhang’s laboratory and RiboBio Co., Guangzhou began cooperation which is supported by a Science Innovation and Development Grant of the Guangzhou government. The project focuses on the development of application using CRISPR/Cas9 gene editing and accessible synthetic chemistry for RNA synthesis in CRISPR/Cas9 editing, so that it progressively builds the world’s first human genome library which constitutes a scientific and reliable system of CRISPR/Cas9 high-throughput screening analysis, to support the mass application and industrialisation of CRISPR/Cas9 technology. Zhang’s laboratory is responsible for developing data analysis platform for high-throughput screening with CRISPR/Cas9 editing. More specifically, our laboratory employs and improves the early high-throughput screening system with siRNA library, and designs mathematical models for target library, reference setup, microplate selection and sample quantification compliant with CRISPR/Cas9 editing. Also, we introduce quality control on data in experimental design to reduce systemic errors.
Source: UMagazine Issue 20